Influenza is ubiquitous. Every fall, we line up to get our flu shots with the hope that we will be protected from the virus that infects 10 to 20 percent of people worldwide each year. But some years, the vaccine is less effective than others.
Every year, CDC scientists engineer a new flu virus. By examining phylogenetic relationships, which are based on shared common ancestry and relatedness, researchers identify virus strains to target with a vaccine for the following flu season.
Sometimes, they do a good job predicting which strains will flourish in the upcoming flu season; other times, they pick wrong.
Andrew Pekosz, PhD, is a researcher at Johns Hopkins who examines why we fail to predict strains to target with vaccines. In particular, he examines years when the vaccine was ineffective and the viruses that were most prevalent to identify properties of these strains.
A virus consists of RNA enclosed in a membrane. Vaccines function by targeting membrane proteins that facilitate movement of the viral genome into host cells that it is infecting. For the flu virus, this protein is hemagglutinin (HA). An additional membrane protein called neuraminidase (NA) allows the virus to release itself from a cell it has infected and prevents it from returning to infected cells.
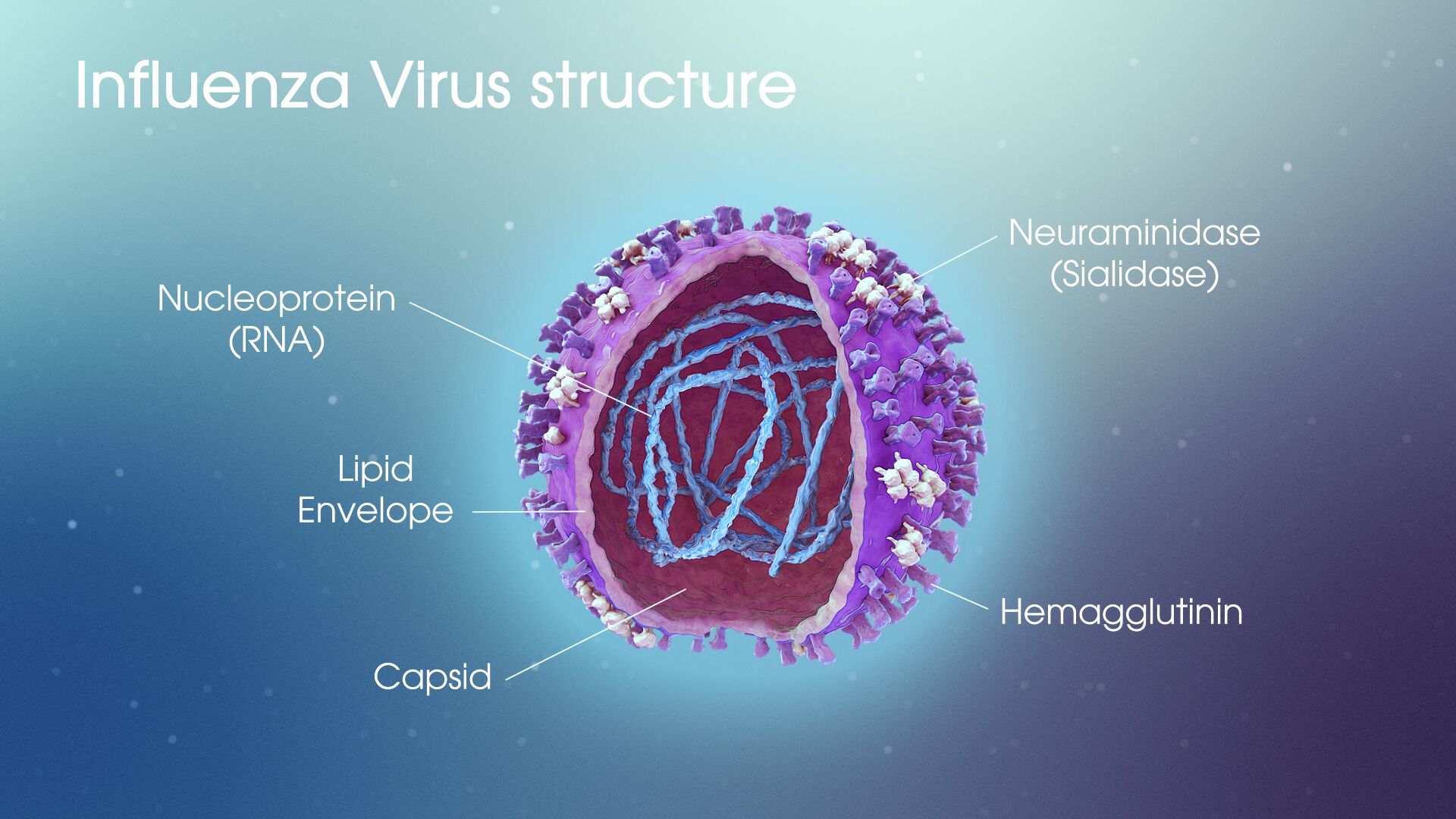
Studying the viruses that flourished in the 2014-2015 and 2016-2017 flu seasons, Pekosz and his team have identified mutations to these surface proteins that allowed certain strains to evade the vaccine.
In the 2014-2015 season, a mutation in the HA receptor conferred an advantage to the virus, but only in the presence of the antibodies present in the vaccine. In the absence of these antibodies, this mutation was actually detrimental to the virus’s fitness. The strain was present in low numbers in the beginning of the flu season, but the selective pressure of the vaccine pushed it to become the dominant strain by the end.
The 2016-2017 flu season saw a similar pattern of mutation, but in the NA protein. The part of the virus membrane where the antibody binds, or the epitope, was covered in the mutated viral strain. Since the antibodies produced in response to the vaccine could not effectively identify the virus, the vaccine was ineffective for these mutated strains.
With the speed at which the flu virus evolves, and the fact that numerous strains can be active in any given flu season, engineering an effective vaccine is daunting. Pekosz’s findings on how these vaccines have previously failed will likely prove invaluable at combating such a persistent and common public health concern.

