DNA extracted from a 1,475-year-old jawbone reveals genetic blueprint for one of the largest lemurs ever.
If you’ve been to the Duke Lemur Center, perhaps you’ve seen these cute mouse- to cat-sized primates leaping through the trees. Now imagine a lemur as big as a gorilla, lumbering its way through the forest as it munches on leaves.
It may sound like a scene from a science fiction thriller, but from skeletal remains we know that at least 17 supersized lemurs once roamed the African island of Madagascar. All of them were two to 20 times heftier than the average lemur living today, some weighing up to 350 pounds.
Then, sometime after humans arrived on the island, these creatures started disappearing.
The reasons for their extinction remain a mystery, but by 500 years ago all of them had vanished.
Coaxing molecular clues to their lives from the bones and teeth they left behind has proved a struggle, because after all this time their DNA is so degraded.
But now, thanks to advances in our ability to read ancient DNA, a giant lemur that may have fallen into a cave or sinkhole near the island’s southern coast nearly 1,500 years ago has had much of its DNA pieced together again. Researchers believe it was a slow-moving 200-pound vegetarian with a pig-like snout, long arms, and powerful grasping feet for hanging upside down from branches.
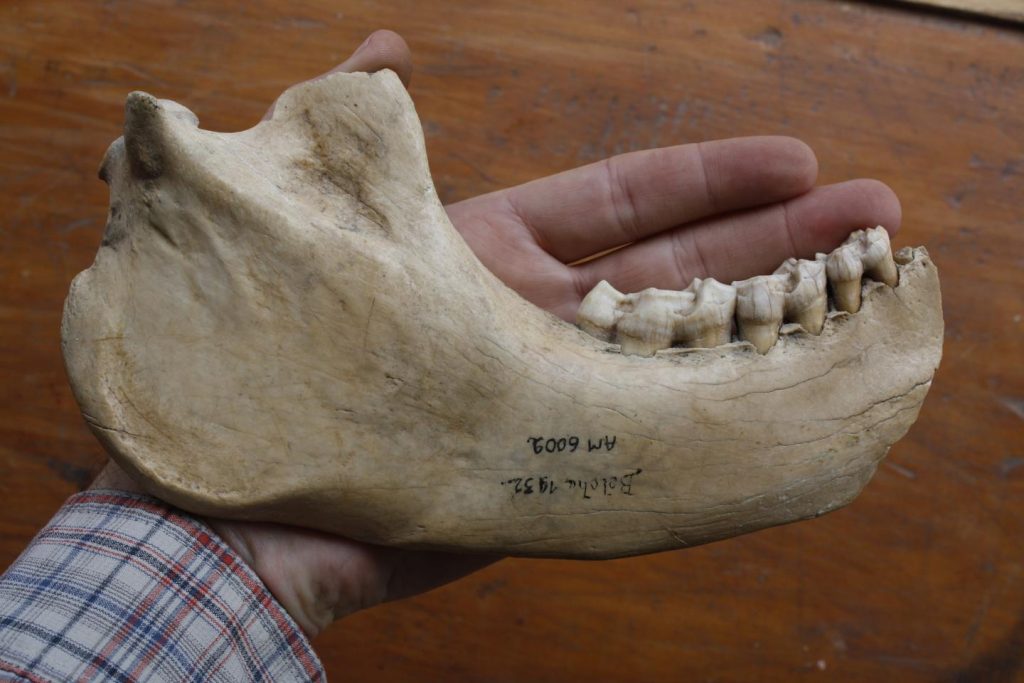
A single jawbone, stored at Madagascar’s University of Antananarivo, was all the researchers had. But that contained enough traces of DNA for a team led by George Perry and Stephanie Marciniak at Penn State to reconstruct the nuclear genome for one of the largest giant lemurs, Megaladapis edwardsi, a koala lemur from Madagascar.
Ancient DNA can tell stories about species that have long since vanished, such as how they lived and what they were related to. But sequencing DNA from partially fossilized remains is no small feat, because DNA breaks down over time. And because the DNA is no longer intact, researchers have to take these fragments and figure out their correct order, like the pieces of a mystery jigsaw puzzle with no image on the box.

Hard-won history lessons
The first genetic study of M. edwardsi, published in 2005 by Duke’s Anne Yoder, was based on DNA stored not in the nucleus — which houses most of our genes — but in another cellular compartment called the mitochondria that has its own genetic material. Mitochondria are plentiful in animal cells, which makes it easier to find their DNA.
At the time, ancient DNA researchers considered themselves lucky to get just a few hundred letters of an extinct animal’s genetic code. In the latest study they managed to tease out and reconstruct some one million of them.
“I never even dreamed that the day would come that we could produce whole genomes,” said Yoder, who has been studying ancient DNA in extinct lemurs for over 20 years and is a co-author of the current paper.
For the latest study, the researchers tried to extract DNA from hundreds of giant lemur specimens, but only one yielded enough useful material to reconstitute the whole genome.
Once the creature’s genome was sequenced, the team was able to compare it to the genomes of 47 other living vertebrate species, including five modern lemurs, to identify its closest living relatives. Its genetic similarities with other herbivores suggest it was well adapted for grazing on leaves.
Despite their nickname, koala lemurs weren’t even remotely related to koalas. Their DNA confirms that they belonged to the same evolutionary lineage as lemurs living today.
To Yoder it’s another piece of evidence that the ancestors of today’s lemurs colonized Madagascar in a single wave.
Since the first ancient DNA studies were published, in the 1980s, scientists have unveiled complete nuclear genomes for other long-lost species, including the woolly mammoth, the passenger pigeon, and even extinct human relatives such as Neanderthals.
Most of these species lived in cooler, drier climates where ancient DNA is better preserved. But this study extends the possibilities of ancient DNA research for our distant primate relatives that lived in the tropics, where exposure to heat, sunlight and humidity can cause DNA to break down faster.
“Tropical conditions are death to DNA,” Yoder said. “It’s so exciting to get a deeper glimpse into what these animals were doing and have that validated and verified.”
See them for yourself
Assembled in drawers and cabinets cases in the Duke Lemur Center’s Division of Fossil Primates on Broad St. are the remains of at least eight species of giant lemurs that you can no longer find in the wild. If you live in Durham, you may drive by them every day and have no idea. It’s the world’s largest collection.
In one case are partially fossilized bits of jaws, skulls and leg bones from Madagascar’s extinct koala lemurs. Nearby are the remains of the monkey-like Archaeolemur edwardsi, which was once widespread across the island. There’s even a complete skeleton of a sloth lemur that would have weighed in at nearly 80 pounds, Palaeopropithecus kelyus, hanging upside down from a branch.
Most of these specimens were collected over 25 years between 1983 and 2008, when Duke Lemur Center teams went to Madagascar to collect fossils from caves and ancient swamps across the island.
“What is really exciting about getting better and better genetic data from the subfossils, is we may discover more genetically distinct species than only the fossil record can reveal,” said Duke paleontologist Matt Borths, who curates the collection. “That in turn may help us better understand how many species were lost in the recent past.”
They plan to return in 2022. “Hopefully there is more Megaladapis to discover,” Borths said.

CITATION: “Evolutionary and Phylogenetic Insights From a Nuclear Genome Sequence of the Extinct, Giant, ‘Subfossil’ Koala Lemur Megaladapis Edwardsi,” Stephanie Marciniak, Mehreen R. Mughal, Laurie R. Godfrey, Richard J. Bankoff, Heritiana Randrianatoandro, Brooke E. Crowley, Christina M. Bergey, Kathleen M. Muldoon, Jeannot Randrianasy, Brigitte M. Raharivololona, Stephan C. Schuster, Ripan S. Malhi, Anne D. Yoder, Edward E. Louis Jr, Logan Kistler, and George H. Perry. PNAS, June 29, 2021. DOI: 10.1073/pnas.2022117118.