By David Jarmul
For years, scientists looking to develop better antibiotics have had their eye on an enzyme that lies just within the membrane of potentially deadly bacteria. The enzyme, MraY, plays a critical role in the process by which the bacteria build the walls of their own cells as they develop.
Penicillin and related drugs such as amoxicillin block this process, thereby preventing the bacteria from going on to infect humans with everything from strep throats to ear infections. Scientists around the world have been working to improve or replace these drugs, especially as bacteria develop resistance to them. But progress has been slow, in part because researchers have lacked a clear view of MraY’s shape and how exactly it works.
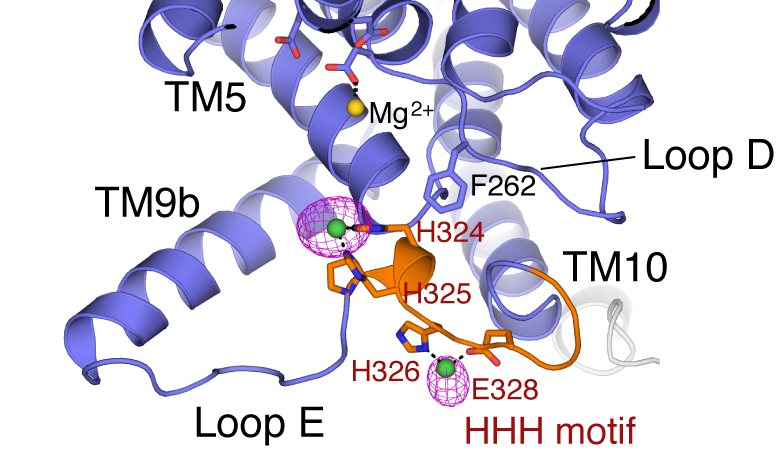
Now the enzyme has come into clearer view. Writing in Science, Seok-Yong Lee and his colleagues at Duke University School of Medicine have published the crystal structure of MraY, which they describe as “an essential membrane enzyme for bacterial cell wall synthesis.”
The structure, at a resolution of 3.3 angstroms, provides the most detailed picture yet of the enzyme’s architecture. It offers new insights to guide drug developers who seek to expand the clinical options available to physicians and their patients. The image below represents a key part of the structure.
Lee, a researcher in Duke’s biochemistry department, and his team worked with colleagues to analyze data they gathered at the Advanced Photon Source at Argonne National Laboratory. Their research was supported primarily with start-up funds from the Duke University Medical Center.
CITATION: “Crystal Structure of MraY, an Essential Membrane Enzyme for Bacterial Cell Wall Synthesis,” Ben C. Chung, Jinshi Zhao, Robert A. Gillespie, Do-Yeon Kwon, Ziqiang Guan, Jiyong Hong, Pei Zhou, Seok-Yong Lee. Science, Aug. 30, 2013.